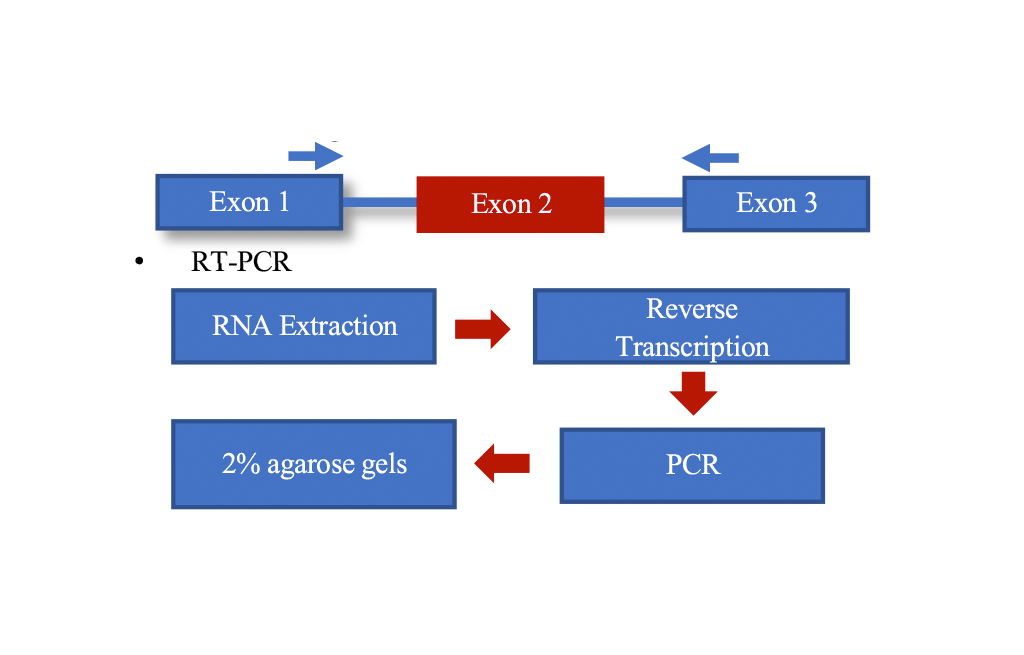
Ribonucleic acids (RNAs) in cells are associated with RNA-binding proteins (RBPs), and this project works with a specific type of RBPs, called heterogeneous nuclear ribonucleoproteins (hnRNPs), which contribute to multiple aspects of RNA metabolism, including alternative splicing, which is the process during gene expression, where a single gene is expressed as multiple isoforms that code for different proteins. This project investigates whether the genes that encode the hnRNPs are alternatively spliced, such that the regulators of splicing (i.e. hnRNPs) are themselves regulated at the splicing level. Alternative splicing of a given hnRNP would generate different versions of that hnRNP with various potential functions with regards to the alternative splicing of its targets, which could lead to diseases, such as cancer. This has not been extensively studied before, especially in the context of breast cancer. This project works with a panel of 12 hnRNPs, studying their alternative splicing patterns, primarily using RT-PCR and bioinformatics, and this is done on breast cancer cells as breast cancer is the most common cancer in Qatar. In addition to studying the alternative splicing patterns of these 12 hnRNPs, this project also narrows down the hnRNPs to one, which is hnRNPA2B1 based on many factors, and this hnRNP is further studied computationally. The ultimate aim of this research is to uncover breast cancer-specific alternative splicing of these hnRNPs, which could set the foundation for novel therapeutic approaches as the expression level of hnRNPs is altered in many types of cancer, suggesting their role in tumorigenesis.